|
|
Ming Xiao, PhD
Professor
School of Biomedical Engineering, Science and Health Systems
Office: Bossone 210
Phone: 215.895.2690
Email:
mx44@drexel.edu
|
Bio
Dr. Xiao’s research focuses on developing genomic technologies and applying them in studying the human genome. Among his recent work at Drexel is the development of technologies based on the CRISPR-Cas9 system for genome mapping and sequencing, which have broad applications in molecular diagnostics, cell/gene therapies, and faster cancer screening. Using this and other technologies, they have gained many significant insights into human genome structures and variations.
Education
- PhD, Biophysics, Baylor University, 1997
- BS, Biomedical Engineering, Huazhong University of Science and Technology, 1988
Research Interests
Micro/nanotechnology, single-molecule detection, genomic technology, DNA/RNA chemistry, human genomics, and bioinformatics.
Publications
The publications listed here are the selected publications and not the full list.
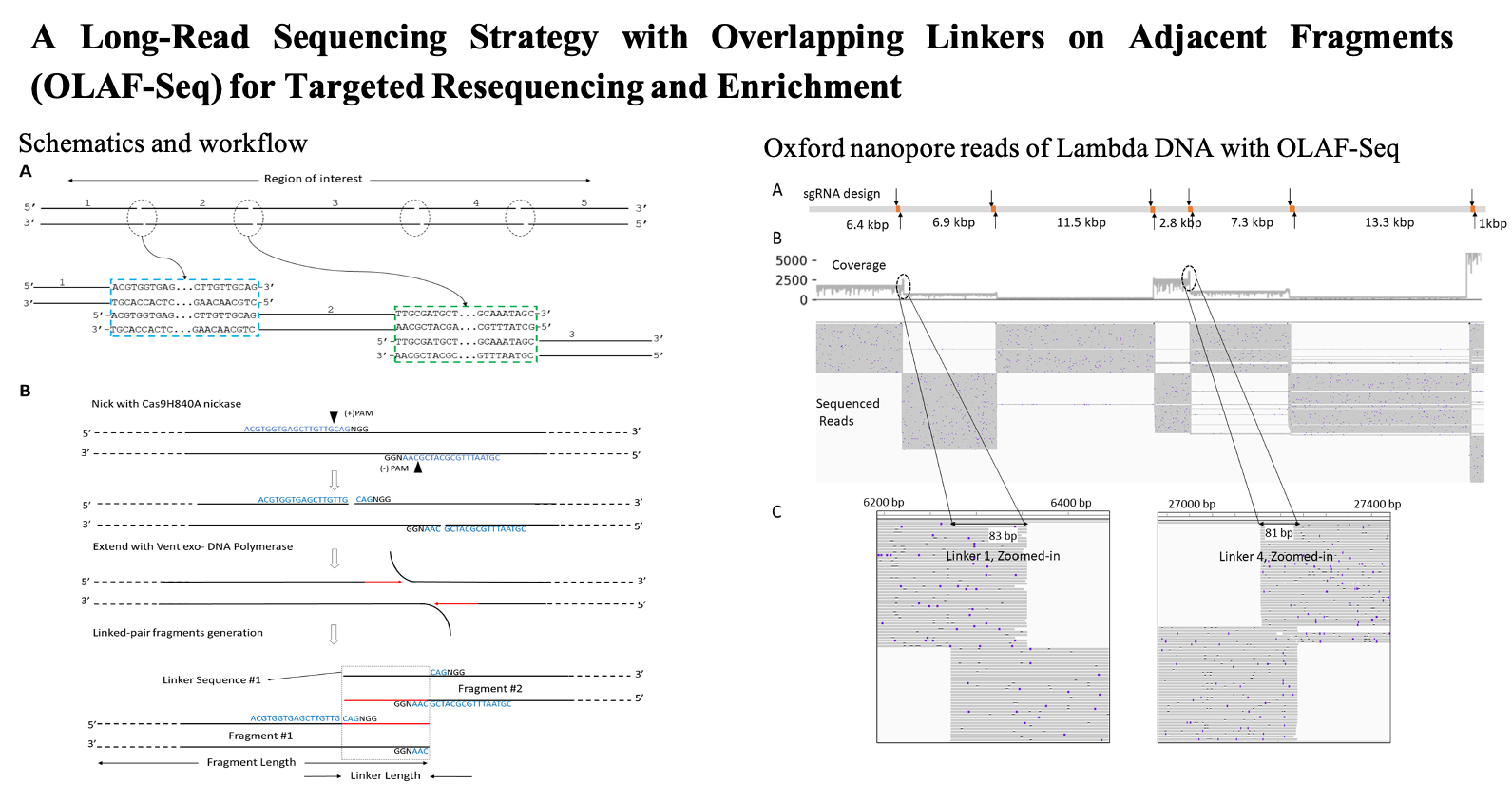
- Uppuluri, L., Wang, Y., K., Y., Mell, J. and Xiao, M. (2024) A Long-Read Sequencing Strategy with Overlapping Linkers on Adjacent Fragments (OLAF-Seq) for Targeted Resequencing and Enrichment (in press).
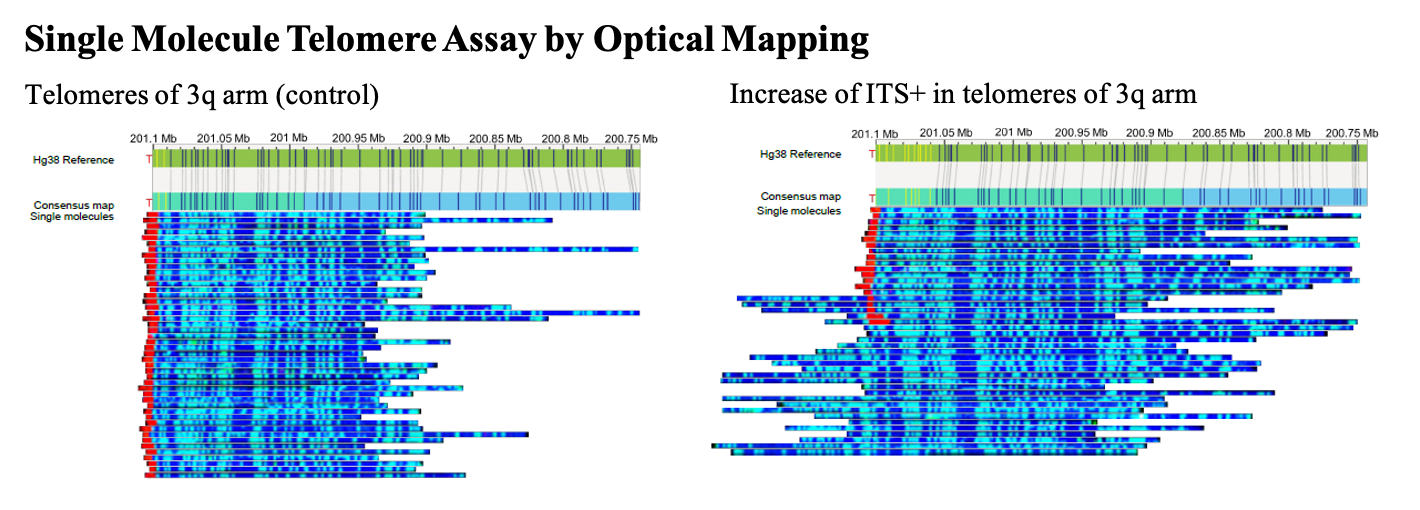
- Raseley, K., et al., Single-Molecule Telomere Assay via Optical Mapping (SMTA-OM) Can Potentially Define the ALT Positivity of Cancer. Genes, 2023. 14(6): p. 1278.
- Uppuluri, L., Jahave, T., Wang, Y. and Xiao, M. (2021) Multiple-color whole-genome mapping in nanochannel for genetic analysis. Analytical Chemistry 93 (28), 9808-9816.
- Abid, H.Z., Young, E., McCaffrey, J., Raseley, K., Varapula, D., Wang, H.-Y., Piazza, D., Mell, J. and Xiao, M. (2021) Customized optical mapping by CRISPR–Cas9 mediated DNA labeling with multiple sgRNAs. Nucleic Acids Research, 49, e8-e8.
- Young, E., Abid, H.Z., Kwok, P.-Y., Riethman, H. and Xiao, M. (2020) Comprehensive Analysis of Human Subtelomeres by Whole Genome Mapping. PLOS Genetics, 16, e1008347.
- Wong, K.H.Y., Ma, W., Wei, C.-Y., Yeh, E.-C., Lin, W.-J., Wang, E.H.F., Su, J.-P., Hsieh, F.-J., Kao, H.-J., Chen, H.-H. et al. (2020) Towards a reference genome that captures global genetic diversity. Nature Communications, 11, 5482.
- Varapula, D., LaBouff, E., Raseley, K., Uppuluri, L., Ehrlich, G.D., Noh, M. and Xiao, M. (2019) A micropatterned substrate for on-surface enzymatic labeling of linearized long DNA molecules. Scientific Reports, 9, 15059.
- Levy-Sakin, M., Pastor, S., Mostovoy, Y., Li, L., Leung, A.K.Y., McCaffrey, J., Young, E., Lam, E.T., Hastie, A.R., Wong, K.H.Y., et al. (2019) Genome maps across 26 human populations reveal population-specific patterns of structural variation. Nature Communications, 10, 1025.
- McCaffrey, J., Young, E., Lassahn, K., Sibert, J., Pastor, S., Riethman, H., and Xiao, M. (2017) High-throughput single-molecule telomere characterization. Genome Research, 27, 1904-1915.
- McCaffrey, J., Sibert, J., Zhang, B., Zhang, Y., Hu, W., Riethman, H. and Xiao, M. (2015) CRISPR-CAS9 D10A nickase target-specific fluorescent labeling of double-strand DNA for whole genome mapping and structural variation analysis. Nucleic Acids Research, 44, e11-e11.
- Lam, E.T., Hastie, A., Lin, C., Ehrlich, D., Das, S.K., Austin, M.D., Deshpande, P., Cao, H., Nagarajan, N., Xiao, M. et al. (2012) Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nature Biotechnology, 30, 771-776.
- Baday, M., Cravens, A., Hastie, A., Kim, H., Kudeki, D.E., Kwok, P.-Y., Xiao, M. and Selvin, P.R. (2012) Multicolor Super-Resolution DNA Imaging for Genetic Analysis. Nano Letters, 12, 3861-3866.
- Das, S.K., Austin, M.D., Akana, M.C., Deshpande, P., Cao, H. and Xiao, M. (2010) Single-molecule linear analysis of DNA in nano-channel labeled with sequence-specific fluorescent probes. Nucleic Acids Research, 38, e177-e177.
- Xiao, M., Wan, E., Chu, C., Hsueh, W.-C., Cao, Y. and Kwok, P.-Y. (2009) Direct determination of haplotypes from single DNA molecules. Nature Methods, 6, 199-201.
- Xiao, M., Phong, A., Ha, C., Chan, T.-F., Cai, D., Leung, L., Wan, E., Kistler, A.L., DeRisi, J.L., Selvin, P.R. et al. (2007) Rapid DNA mapping by fluorescent single molecule detection. Nucleic Acids Research, 35, e16-e16.
- Frazer, K.A., Ballinger, D.G., Cox, D.R., Hinds, D.A., Stuve, L.L., Gibbs, R.A., Belmont, J.W., Boudreau, A., Hardenbol, P., Leal, S.M. et al. (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature, 449, 851-861.
- Xiao, M., Reifenberger, J.G., Wells, A.L., Baldacchino, C., Chen, L.-Q., Ge, P., Sweeney, H.L. and Selvin, P.R. (2003) An actin-dependent conformational change in myosin. Nature Structural & Molecular Biology, 10, 402-408.
- Xiao, M. and Selvin, P.R. (2001) Quantum Yields of Luminescent Lanthanide Chelates and Far-Red Dyes Measured by Resonance Energy Transfer. Journal of the American Chemical Society, 123, 7067-7073.
- Xiao, M., Li, H., Snyder, G.E., Cooke, R., Yount, R.G. and Selvin, P.R. (1998) Conformational changes between the active-site and regulatory light chain of myosin as determined by luminescence resonance energy transfer: The effect of nucleotides and actin. Proceedings of the National Academy of Sciences, 95, 15309.
Issued Patents
- US10995364B2 (2021) Methods and devices for single-molecule whole genome analysis.
- US10640810B2 (2020) Methods of specifically labeling nucleic acids using CRISPR/Cas9.
- US10435739B2 (2019) Methods and devices for single-molecule whole genome analysis.
- US10000803B2 (2018) Polynucleotide mapping and sequencing.
- US9758780B2 (2017) Whole genome mapping by DNA sequencing with linked-paired-end library.
- US20160046992A1 (2020) Characterization of molecules in nanofluidics.
- EP2954069B1 (2020) EPO Methods for single-molecule analysis.
- US9536041B2 (2017) Methods and devices for single-molecule whole genome analysis.
- US9181578B2 (2015) Polynucleotide mapping and sequencing.
- EP2370594B1 (2014) EPO Polynucleotide mapping and sequencing.
- US8628919B2 (2014) Methods and devices for single-molecule whole genome analysis.
- EP2318547B1 (2018) EPO Methods for single-molecule whole genome analysis.
- EP2664677B1 EPO Methods for single-molecule whole genome analysis.
- US7771944B2 (2010) Methods for determining genetic haplotypes and DNA mapping.
- US7829278B2 (2010) Polynucleotide barcoding.